It’s Sydney Science Festival this week so I’m going to 4 talks and liveblogging them. Tonight’s talk by Simon Ho is Time After Time: Measuring Evolution with Molecular Clocks, discussing the research he’s been involved with.
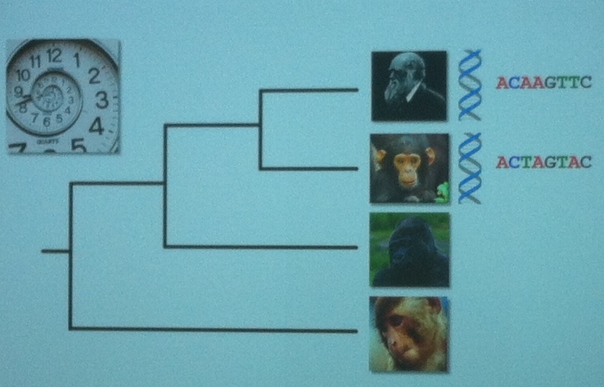
Phylogenetic tree with Darwin, a chimp, gorilla and ape I can’t identify with different genetic code being shown for Darwin and the chimp.
When we look at the picture we’ve built up of the history of life over the last 3.5 billion years, we have increasing diversity of life punctuated by mass extinctions (eg. the Cretaceous extinction that killed the dinosaurs and the much larger Permian extinction). However, the details have mainly been obtained through the fossil record which is systematically incomplete for these reasons:
- Single-celled and small organisms (which made up most of life for most of the time) aren’t preserved well
- These organisms evolve very quickly too so our knowledge is even more incomplete
- Soft-bodied organisms (which made up most of life a long time ago) aren’t preserved well
- Very old periods have few fossils since they have been degraded by geological activity.
Molecular clocks are a way to fill this gap by performing computations on the genome of modern organisms. Or extinct ones, if they are recent and have been refrigerated, eg. mammoths.
The outcome of such a study is an evolutionary tree that shows the relationship between the organisms. The root is the common ancestor and each node is a divergence point. The branches lengths can represent the amount of genetic change (phylogram) or the length of time that the branch has been around (chronogram). Once the diagram is done, we can correlate divergence points to other information (eg. geological events) to learn more about the evolutionary history of a species or group.

Here’s how it’s done in a simplified scenario:
- Genetic change estimate: Two species are compared for the difference in genomes (eg. 1.4% between humans and chimps).
- Calibration point: Their nearest common ancestor is determined and dated through other methods (eg. fossil record).
- This is used to calculate the rate variation or speed at which the genome changes.
These numbers are then applied to other species in the set, with the genomic differences being used to directly estimate their last point of divergence. A supercomputer then comes up with the most likely tree consistent with all the information.
There are complications: for example the rate of genetic change is related to the length of a generation. During one rhino generation, there will be 200 fly generations so flies evolve faster. Studies that span long periods of time need to account for this (and different rates of change on every branch) because the species are very different if you go back in time.
Ho went through a number of studies that he was involved with applying molecular clocks to find out some interesting things:
- A chronogram of hepatitis C looked at their genetic diversity to estimate the number of infections and change over time during the 20th century, then correlate that to changes in public health policy.
- A chronogram of the potato blight virus (that caused the Irish potato famine) helped identify the origins of that particular strain.
- A chronogram of extinct ice age megafauna (and a study of their genetic diversity) was correlated with the fauna’s changing habitats to try determine the extent to which early humans were responsible for the extinction due to hunting. As opposed to climate change. It was a bit of both.
- A chronogram of modern birds showed that most modern bird groups diverged about 65 million years ago or right after the dinosaurs went extinct. This filled the gap in the fossil record and confirmed that birds quickly filled niches left by dinosaurs.
- A chronogram of flowering plants also showed that they diversified into their modern groups much earlier than they actually appear in the fossil record.
- A chronogram of isopods (which includes that disgusting species that becomes the tongue of a fish) showed that they diverged and went into the sea just before the Permian extinction meaning they found habitats that let them survive it.
- A chronogram of insects showed that they diversified before flowering plants and so at first did not substantially co-
Finally molecular clocks are much less reliable for earlier forms of life (eg. Precambrian). This is because after so much time, new mutations start to erase the original genetic mutations and the signal/noise ratio reduces. More sophisticated analysis techniques are needed to make progress with these.
Q: Since species change at different rates, won’t the genome change at different rates?
A: Even if an organism doesn’t change in appearance, the genome changes because of genetic drift. The changes used by the molecular clock can be silent (ie. not expressed by a chance in the organism).
Q: Are there other sources for clocks?
A: Originally it was based on proteins, which is useful for older fossils since the DNA code gets degraded more (since there are only 4 letters). Now it’s mainly DNA.
Q: There is now evidence that some Indigenous Australians and Pacific Islanders got to the Americas as well as those who crossed via the Bering land bridge; can molecular clocks find out who got there first?
A: Maybe – even a single person’s genome provides lots of useful info.
Q: Does an organism’s complexity and/or environmental stress affect the rate of change?
A: Depends on what you mean by complexity. Genome size and number of cell types don’t necessarily correlate with genetic change. But more complex organisms may have longer generations which would slow down their rate of evolution.
Q: Since molecular clocks mostly use DNA from a cells mitochondria (which have much fewer genes than nuclear DNA), isn’t this missing important information?
A: Yes. Mitochondrial DNA is the standard because it’s conserved across different species and every cell has lots of copies so it’s easier to sequence and analyse. But there will be a need to use nuclear DNA more in the future
A great talk; I learned heaps!




0 Comments